Electronic health record clinical and genomic data provides relevant insights for non-small cell lung cancer
1. In patients with non-small cell lung cancer (NSCLC), an analysis of clinical and genomic data from routine care replicated previously established relationships between driver mutations and response to therapy and between tumor mutation burden and response to immunotherapy
2. The study provides confirmation that a multi-institution clinicogenomic database may provide relevant insights in oncology.
Evidence Rating Level: 2 (Good)
Study Rundown: Most clinicogenomic associations in targeted therapeutics for patients with cancer arise from small trials, single institution studies, or national registries. There is a need to better understand the benefit and optimal use of these therapeutics in large populations where the findings can significantly advance clinical practice. The current study evaluated whether a continuously updating database linked to comprehensive genomic profiling (CGP) that included patient data obtained in routine clinical care could yield established associations and relevant insights into patients with non-small cell lung cancer (NSCLC). In those with a driver mutation, improved overall survival was observed in those treated with targeted therapy. Tumor mutation burden (TMB) was significantly higher in smokers and lower in patients with alterations in EGFR, ALK, or ROS1. Further, in patients treated with anti-PD-1/PD-L1 therapies, TMB of 20 ((in mutations/megabase) or more was associated with improved overall survival from treatment initiation.
Overall, this study suggests that datasets linking clinicogenomic profiling to clinical outcomes in NSCLC can provide pertinent findings in patients with NSCLC and indicate the feasibility of a clinicogenomic database from routine patient care as a possible and relevant approach for further research in oncology. This study is limited in its overall survival analysis by the completeness of the underlying mortality data as well as other biases that the study authors have taken efforts to reduce. Further research is required to validate this approach as a useful tool in oncology.
Click to read the study, published today in JAMA
Relevant Reading: Real-World Evidence in Support of Precision Medicine: Clinico-Genomic Cancer Data As A Case Study
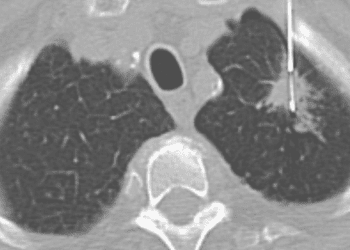
In-Depth [retrospective cohort]: De-identified patient cohort was obtained from linking the Flatiron Health EHR database to the Foundation Medicine database of tumor sequencing results. An NSCLC subcohort was identified using ICD codes and included 4064 patients with NSCLC (median age 66.0 years; 51.9% females; 78.3% with history of smoking; 77.6% with nonsquamous cancer) in the database, of which about 21.4% had an alteration in EGFR, ALK, or ROS1. These patients had to be seen at an oncology practice at least once after January 2011, must have a confirmed diagnosis of NSCLC after manual review of record, and had CGP testing on a tumor specimen that was determined by a pathologist to be NSCLC or tumor of unknown primary. Tumor genotyping, PD-L1 data, and clinical data were obtained from FoundationOne and EHR records. Main end points included overall survival, time receiving therapy, maximal therapeutic response, and clinical benefit rate. In patients with a driver mutation, improved overall survival was observed among those treated with a targeted therapy vs non-targeted therapy (median, 18.6 months vs 11.4 months from advanced diagnosis; p < 0.001). Tumor mutation burden (in mutations/Mb) was higher among smokers vs non-smokers (8.7 vs 2.6, p < 0.001) and lower among among patients with vs without EGFR, ALK, or ROS1 alteration. In patients treated with anti-PD-1/PD-L1 therapies, TMB of 20 or more was more significantly associated with improved overall survival from therapy (16.8 months vs 8.5 months; p < 0.001).
Image: PD
©2019 2 Minute Medicine, Inc. All rights reserved. No works may be reproduced without expressed written consent from 2 Minute Medicine, Inc. Inquire about licensing here. No article should be construed as medical advice and is not intended as such by the authors or by 2 Minute Medicine, Inc.